The FOMO25 Challenge
Overview
For frequently asked questions, please see our FAQ.
This challenge seeks to investigate the few-shot generalisation properties of foundation models in the context of real-world brain MRI data by first pretraining on a large unlabelled dataset before evaluating models on three large clinical, multi-vendor, and multi-center datasets. The concept is to evaluate the same pretrained models on multiple downstream tasks, to understand the effects of different pretraining paradigms and configurations on downstream performance and ultimately both identify the most promising methodologies and quantify the benefits of self-supervised pretraining.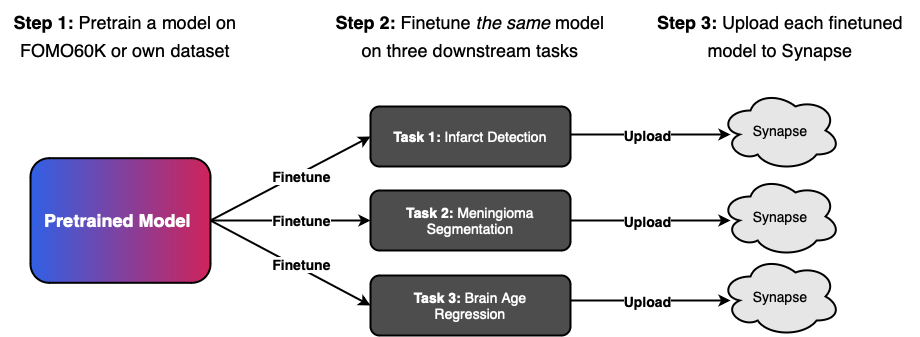
Figure 1: Participants will first pretrain on a large unlabelled dataset before evaluating the same pretrained model on three large clinical, multi-vendor, and multi-center datasets.
Challenge Logistics TL;DR
Code available for both finetuning and pretraining
Participants are provided with a baseline framework to carry out both pretraining and finetuning, enabling them to concentrate on specific components of the workflow. The code is available here.
Cash prices of $2000
Participants will compete for a total cash prize of $1000 for each track, distributed among the top-performing teams. The exact distribution will be announced closer to the competition deadline. Winners will be determined based on their aggregated performance on the evaluation metrics for each task.
Two submission tracks: "Methods" and "Open".
On the "Method" track, participants are restricted to using the provided FOMO60K dataset for pretraining. On the "Open" track, participants can pretrain on any data, including private data. Participants are not allowed to use additional data for finetuning.
Half-day event at MICCAI 2025 🇰🇷
Results will be announced at half-day event at MICCAI 2025, combined with talks by invited speakers. Top-performing teams will be invited to present. The event will be held on 22 September or 28 September. Hope to see you in Daejeon, South Korea!
Timeline
1 April
Challenge opens!
7 April
Code release of pretraining and finetuning code. Available here.
8 May
Access to sanity-check code (technical evaluation which confirms that container is correctly configured). Available here.
15 June
Validation leaderboard opens and final submission pipeline opens. Here.
20 August 31 August, 11.59 p.m. PST (Extended)
Challenge submission deadline.
2 September
Teams invited for presentation will be contacted.
27 September
Half-day event at MICCAI 2025.
Tasks & Data
Pretraining Data
- Subjects: 11,187
- Sessions: 13,900
- Scans: 60,529
- Sequences: T1, T2, Flair, T1c, T2*, GRE, minIP, ADC, SWI, DWI and more.
Finetuning Data
- Infarcts: 21 cases
- Meningiomas: 25 cases
- Brain Age: 200 cases
Evaluation Data
- Infarcts: 400 cases
- Meningiomas: 132 cases
- Brain Age: 1,000 cases
Note: 20% of the evaluation data will be used for pre-evaluation and a pre-deadline leaderboard.
Pretraining
The provided pretraining data is a large-scale dataset of brain MRI scans, including both clinical and research-grade scans. The dataset includes a wide range of sequences, including T1, MPRAGE, T2, T2*, FLAIR, SWI, T1c, PD, DWI, ADC, and more. The dataset is split into 11,167 subjects, with a total of 13,900 sessions and 60,529 scans. All data is provided as NiFTY-files.
The dataset format is provided as one large standardized, preprocessed (including skull stripped, RAS reoriented, co-registered) dataset and is collected from the following public sources: OASIS [1, 2], BraTS [3-7], MSD [8], IXI [9], MGH Wild [10], NKI [11], SOOP [12], NIMH [13], DLBS [14], IDEAS [15], ARC [16], MBSR [17], UCLA [18] QTAB [19], AOMIC ID1000 [20].
Please be aware that usage of the dataset is subject to a Data Usage Agreement and a Citation Policy.
FOMO-60K is available for download via Huggingface.
Baseline models pretrained on FOMO-60K are available for download via Huggingface.
Finetuning
Task 1: Infarct Detection
Image-level binary classification of the presence of infarct(s) in the brain.
Cohort: 18-year-old or older patients with possible brain infarct(s). Subjects underwent an MRI scan and were subsequently diagnosed with infarcts and a control group in 2019 in multiple hospitals in Denmark (evaluation data) and India (finetuning data). Finetuning data is acquired from GE, Siemens scanners. Evaluation data is acquired from a diverse set of scanners from GE, Siemens and Philips. All scanners are 1.5T or 3T.
Sequences: Sequences always include T2 FLAIR, DWI (b-value 1000), ADC, and either T2* or SWI images.
Available additional information: A binary mask of the infarcts is provided for the finetuning data. This mask is not provided for the evaluation data. Note that the masks have not been verified by expert radiologists and are meant to serve as approximate annotations only.
Dataset sizes: Finetune cases: 21. Validation cases: 80. Test cases: 320. Each case represent different subjects.
Assessment Metric: AUROC (Area Under the Receiver Operator Curve (ROC))
Task 2: Meningioma Segmentation
Binary segmentation of brain meningiomas on MRI scans.
Cohort: 18-year-old or older preoperative meningioma patients. Subjects who underwent an MRI diagnosed with preoperative meningioma in 2019 in multiple hospitals in Denmark (evaluation data) and India (finetuning data). Finetuning data is acquired from GE, Siemens scanners. Evaluation data is acquired from a diverse set of scanners from GE, Siemens and Philips. All scanners are 1.5T or 3T.
Sequences: Sequences always include T2 FLAIR, DWI (b-value 1000), and either T2* or SWI images.
Available additional information: For the finetuning data, a binary mask of meningiomas will be provided.
Dataset sizes: Finetuning cases: 23. Validation cases: 40. Test cases: 160. Each case represent different subjects.
Assessment Metric: Overlap-based metric: Dice Similarity Coefficient (DSC). Boundary-based metric: Normal Surface Distance (NSD)
Task 3: Brain Age Regression
Accurate prediction of the age of the patient based on MRI scans.
Cohort: Patients 18 years or older with no underlying brain conditions defined as patients with no neurological conditions and not on brain-related medication. Subjects who underwent an MRI with no underlying brain conditions in 2019 in multiple hospitals in Denmark (evaluation data) and Boston (finetuning data).
Sequences: Sequences always includes T1w and T2w MRI scans.
Available additional information: For each case, age (represented by an integer) at the time of the MRI visit is provided.
Dataset sizes: Finetuning cases: 200. Validation Cases: 200. Test cases: 800 Each case represent different subjects.
Assessment Metric: Absolute Error (AE), Correlation Coefficitent
Code
Participants are provided with a baseline framework to carry out both pretraining and finetuning, enabling participants to concentrate on specific components of the workflow. The provided code is designed to be a starting point, and not required for participation. The baseline framework includes scripts for data preprocessing, model training, and evaluation. It is designed to be modular and extensible, allowing participants to easily integrate their own methods. Detailed documentation and examples are provided in the repository to help participants get started quickly. The code is available here.
Pretraining
The pretraining code is based on Masked Autoencoders (MAE) using the AMAES framework [21]. This framework provides a resilient and efficient implementation for self-supervised learning on medical imaging data, enabling participants to leverage state-of-the-art techniques for pretraining.
Finetuning
The finetuning code is using the Yucca framework [22]. The framework is designed to simplify the process of finetuning models for medical image analysis tasks, providing utilities for data handling, model training, and evaluation.
Leaderboard & Evaluation
Tracks
For all tasks, the challenge will have two tracks:
- Methods Track: No additional data allowed. Challenge participants can only use the large-scale FOMO60K dataset. This challenge track is intended to provide insights into methods development for pretraining, finetuning and model architectures.
- Open Track: Any data allowed for pretraining. Challenge participants can use any data, including private data, for their submissions. This track includes submissions using any form of segmentation maps obtained through manual labor or disease diagnosis information during pretraining. All data used must be specified and properly cited. The track is intended to allow participants to showcase and compare brain MRI Foundation Models.
Leaderboard
For each task, all test cases are assigned uniform weight and each metric is averaged over all cases in the test set.
Per-task Leaderboard
A summary statistic defined as the average of all metrics in the task is used to make a per-task rank. Unified leaderboard: For each pretraining configuration, the same pretrained checkpoint can be finetuned to each of the three tasks. Per pretraining checkpoint, an overall rank is then calculated as the average rank of each task. For partial submissions, that is submissions that have only submitted models for a subset of the three tasks, the worst possible rank is used in the average (defined as number of valid submissions per task plus one.)
Unified Leaderboard
For each pretraining configuration, the same pretrained checkpoint can be finetuned to each of the three tasks. Per pretraining checkpoint, an overall rank is then calculated as the average rank of each task. For partial submissions, that is submissions that have only submitted models for a subset of the three tasks, the worst possible rank is used in the average (defined as number of valid submissions per task plus one.)
Publication Policy
The aim is a publication in a journal such as Nature Methods, Medical Image Analysis, npj Digital Medicine or IEEE Transactions on Medical Imaging. Expected journal submission Q4 2025.
All participating teams that have made substantial submissions (e.g. excludes trivial approaches such as setting all outputs to zero, random guessing, or simply using provided baselines) of a pretrained model to all three tasks will be invited to contribute to the manuscript of the following publication. Each team can have at most five authors on the publication. We will consider exemptions on a case-by-case basis.
Participation Policy
Due to the large number of members in the institues of the organizers, these are divided into two groups: 1. Participants who have active working relations with any of the organizers are not allowed to participate and will not be listed on the leaderboard. 2. Other members of the institutes may participate, but are not eligible for awards and listed with a clearly visible disclaimer in leaderboard.
Cash Price
$1000 for each track, distributed among the top-performing teams. The exact distribution will be announced closer to the competition deadline. Winners will be determined based on the best ranking on the unified leaderboard in each track. We will also seek to raise funding for per-task prizes and for the other track.
Join the Challenge
Be part of the FOMO challenge and contribute to advancing brain MRI research!
References
[1] Open Access Series of Imaging Studies (OASIS): Cross-Sectional MRI Data in Young, Middle Aged, Nondemented, and Demented Older Adults. Marcus, DS, Wang, TH, Parker, J, Csernansky, JG, Morris, JC, Buckner, RL. Journal of Cognitive Neuroscience, 19, 1498-1507.
[2] Open Access Series of Imaging Studies (OASIS): Longitudinal MRI Data in Nondemented and Demented Older Adults. Marcus, DS, Fotenos, AF, Csernansky, JG, Morris, JC, Buckner, RL, 2010. Journal of Cognitive Neuroscience, 22, 2677-2684. doi: 10.1162/jocn.2009.21407
[3] LaBella D, Adewole M, Alonso-Basanta M, Altes T, Anwar SM, Baid U, Bergquist T, Bhalerao R, Chen S, Chung V, Conte GM, Dako F, Eddy J, Ezhov I, Godfrey D, Hilal F, Familiar A, Farahani K, Iglesias JE, Jiang Z, Johanson E, Kazerooni AF, Kent C, Kirkpatrick J, Kofler F, Leemput KV, Li HB, Liu X, Mahtabfar A, McBurney-Lin S, McLean R, Meier Z, Moawad AW, Mongan J, Nedelec P, Pajot M, Piraud M, Rashid A, Reitman Z, Shinohara RT, Velichko Y, Wang C, Warman P, Wiggins W, Aboian M, Albrecht J, Anazodo U, Bakas S, Flanders A, Janas A, Khanna G, Linguraru MG, Menze B, Nada A, Rauschecker AM, Rudie J, Tahon NH, Villanueva-Meyer J, Wiestler B, Calabrese E. The ASNR-MICCAI Brain Tumor Segmentation (BraTS) Challenge 2023: Intracranial Meningioma. ArXiv [Preprint]. 2023 May 12:arXiv:2305.07642v1. PMID: 37608937; PMCID: PMC10441446.
[4] Moawad AW, Janas A, Baid U, Ramakrishnan D, Saluja R, Ashraf N, Maleki N, Jekel L, Yordanov N, Fehringer P, Gkampenis A, Amiruddin R, Manteghinejad A, Adewole M, Albrecht J, Anazodo U, Aneja S, Anwar SM, Bergquist T, Chiang V, Chung V, Conte GM, Dako F, Eddy J, Ezhov I, Khalili N, Farahani K, Iglesias JE, Jiang Z, Johanson E, Kazerooni AF, Kofler F, Krantchev K, LaBella D, Van Leemput K, Li HB, Linguraru MG, Liu X, Meier Z, Menze BH, Moy H, Osenberg K, Piraud M, Reitman Z, Shinohara RT, Wang C, Wiestler B, Wiggins W, Shafique U, Willms K, Avesta A, Bousabarah K, Chakrabarty S, Gennaro N, Holler W, Kaur M, LaMontagne P, Lin M, Lost J, Marcus DS, Maresca R, Merkaj S, Cassinelli Pedersen G, von Reppert M, Sotiras A, Teytelboym O, Tillmans N, Westerhoff M, Youssef A, Godfrey D, Floyd S, Rauschecker A, Villanueva-Meyer J, Pflüger I, Cho J, Bendszus M, Brugnara G, Cramer J, Perez-Carillo GJG, Johnson DR, Kam A, Kwan BYM, Lai L, Lall NU, Memon F, Krycia M, Patro SN, Petrovic B, So TY, Thompson G, Wu L, Schrickel EB, Bansal A, Barkhof F, Besada C, Chu S, Druzgal J, Dusoi A, Farage L, Feltrin F, Fong A, Fung SH, Gray RI, Ikuta I, Iv M, Postma AA, Mahajan A, Joyner D, Krumpelman C, Letourneau-Guillon L, Lincoln CM, Maros ME, Miller E, Morón FEA, Nimchinsky EA, Ozsarlak O, Patel U, Rohatgi S, Saha A, Sayah A, Schwartz ED, Shih R, Shiroishi MS, Small JE, Tanwar M, Valerie J, Weinberg BD, White ML, Young R, Zohrabian VM, Azizova A, Brüßeler MMT, Ghonim M, Ghonim M, Okar A, Pasquini L, Sharifi Y, Singh G, Sollmann N, Soumala T, Taherzadeh M, Vollmuth P, Foltyn-Dumitru M, Malhotra A, Abayazeed AH, Dellepiane F, Lohmann P, Pérez-García VM, Elhalawani H, de Verdier MC, Al-Rubaiey S, Armindo RD, Ashraf K, Asla MM, Badawy M, Bisschop J, Lomer NB, Bukatz J, Chen J, Cimflova P, Corr F, Crawley A, Deptula L, Elakhdar T, Shawali IH, Faghani S, Frick A, Gulati V, Haider MA, Hierro F, Dahl RH, Jacobs SM, Hsieh KJ, Kandemirli SG, Kersting K, Kida L, Kollia S, Koukoulithras I, Li X, Abouelatta A, Mansour A, Maria-Zamfirescu RC, Marsiglia M, Mateo-Camacho YS, McArthur M, McDonnell O, McHugh M, Moassefi M, Morsi SM, Munteanu A, Nandolia KK, Naqvi SR, Nikanpour Y, Alnoury M, Nouh AMA, Pappafava F, Patel MD, Petrucci S, Rawie E, Raymond S, Roohani B, Sabouhi S, Sanchez-Garcia LM, Shaked Z, Suthar PP, Altes T, Isufi E, Dhemesh Y, Gass J, Thacker J, Tarabishy AR, Turner B, Vacca S, Vilanilam GK, Warren D, Weiss D, Worede F, Yousry S, Lerebo W, Aristizabal A, Karargyris A, Kassem H, Pati S, Sheller M, Link KEE, Calabrese E, Tahon NH, Nada A, Velichko YS, Bakas S, Rudie JD, Aboian M. The Brain Tumor Segmentation - Metastases (BraTS-METS) Challenge 2023: Brain Metastasis Segmentation on Pre-treatment MRI. ArXiv [Preprint]. 2024 Dec 9:arXiv:2306.00838v3. PMID: 37396600; PMCID: PMC10312806.
[5] Kazerooni AF, Khalili N, Liu X, Haldar D, Jiang Z, Anwar SM, Albrecht J, Adewole M, Anazodo U, Anderson H, Bagheri S, Baid U, Bergquist T, Borja AJ, Calabrese E, Chung V, Conte GM, Dako F, Eddy J, Ezhov I, Familiar A, Farahani K, Haldar S, Iglesias JE, Janas A, Johansen E, Jones BV, Kofler F, LaBella D, Lai HA, Van Leemput K, Li HB, Maleki N, McAllister AS, Meier Z, Menze B, Moawad AW, Nandolia KK, Pavaine J, Piraud M, Poussaint T, Prabhu SP, Reitman Z, Rodriguez A, Rudie JD, Shaikh IS, Shah LM, Sheth N, Shinohara RT, Tu W, Viswanathan K, Wang C, Ware JB, Wiestler B, Wiggins W, Zapaishchykova A, Aboian M, Bornhorst M, de Blank P, Deutsch M, Fouladi M, Hoffman L, Kann B, Lazow M, Mikael L, Nabavizadeh A, Packer R, Resnick A, Rood B, Vossough A, Bakas S, Linguraru MG. The Brain Tumor Segmentation (BraTS) Challenge 2023: Focus on Pediatrics (CBTN-CONNECT-DIPGR-ASNR-MICCAI BraTS-PEDs). ArXiv [Preprint]. 2024 May 23:arXiv:2305.17033v7. PMID: 37292481; PMCID: PMC10246083.
[6] Correia de Verdier, Maria & Saluja, Rachit & Gagnon, Louis & LaBella, Dominic & Baid, Ujjwall & Tahon, Nourel Hoda & Foltyn-Dumitru, Martha & Zhang, Jikai & Alafif, Maram & Baig, Saif & Chang, Ken & D'Anna, Gennaro & Deptula, Lisa & Gupta, Diviya & Haider, Muhammad Ammar & Hussain, Ali & Iv, Michael & Kontzialis, Marinos & Manning, Paul & Rudie, Jeffrey. (2024). The 2024 Brain Tumor Segmentation (BraTS) challenge: glioma segmentation on post-treatment MRI. 10.48550/arXiv.2405.18368.
[7] Adewole M, Rudie JD, Gbdamosi A, Toyobo O, Raymond C, Zhang D, Omidiji O, Akinola R, Suwaid MA, Emegoakor A, Ojo N, Aguh K, Kalaiwo C, Babatunde G, Ogunleye A, Gbadamosi Y, Iorpagher K, Calabrese E, Aboian M, Linguraru M, Albrecht J, Wiestler B, Kofler F, Janas A, LaBella D, Kzerooni AF, Li HB, Iglesias JE, Farahani K, Eddy J, Bergquist T, Chung V, Shinohara RT, Wiggins W, Reitman Z, Wang C, Liu X, Jiang Z, Familiar A, Van Leemput K, Bukas C, Piraud M, Conte GM, Johansson E, Meier Z, Menze BH, Baid U, Bakas S, Dako F, Fatade A, Anazodo UC. The Brain Tumor Segmentation (BraTS) Challenge 2023: Glioma Segmentation in Sub-Saharan Africa Patient Population (BraTS-Africa). ArXiv [Preprint]. 2023 May 30:arXiv:2305.19369v1. PMID: 37396608; PMCID: PMC10312814.
[8] Simpson, A., Antonelli, M., Bakas, S., Bilello, M., Farahani, K., Ginneken, B., Kopp-Schneider, A., Landman, B., Litjens, G., Menze, B., Ronneberger, O., Sum- mers, R., Bilic, P., Christ, P., Do, R., Gollub, M., Golia-Pernicka, J., Heckers, S.,Jarnagin, W., Cardoso, M.J.: A large annotated medical image dataset for the development and evaluation of segmentation algorithms (02 2019).
[9] http://brain-development.org/ixi-dataset/
[10] Juan E. Iglesias et al. ,SynthSR: A public AI tool to turn heterogeneous clinical brain scans into high-resolution T1-weighted images for 3D morphometry.Sci. Adv.9,eadd3607(2023).DOI:10.1126/sciadv.add3607.
[11] Tobe et al, (2022). A longitudinal resource for studying connectome development and its psychiatric associations during childhood. Scientific Data 9, 300.
[12] Chris Rorden and John Absher and Roger Newman-Norlund (2024). Stroke Outcome Optimization Project (SOOP). OpenNeuro. [Dataset] doi: doi:10.18112/openneuro.ds004889.v1.1.2
[13] A llison C. Nugent and Adam G Thomas and Margaret Mahoney and Alison Gibbons and Jarrod Smith and Antoinette Charles and Jacob S Shaw and Jeffrey D Stout and Anna M Namyst and Arshitha Basavaraj and Eric Earl and Dustin Moraczewski and Emily Guinee and Michael Liu and Travis Riddle and Joseph Snow and Shruti Japee and Morgan Andrews and Adriana Pavletic and Stephen Sinclair and Vinai Roopchansingh and Peter A Bandettini and Joyce Chung (2025). The NIMH Healthy Research Volunteer Dataset. OpenNeuro. [Dataset] doi: doi:10.18112/openneuro.ds005752.v2.1.0
[14] Denise Park and Joseph Hennessee and Evan T. Smith and Micaela Chan and Carson Katen and Julia Bacci and Sarah Frank and Sarah Monier and Alexandra Collyer and Carol Tamminga and William Moore and Neil Rofsky and Karen Rodrigue and Kristen Kennedy and Gagan Wig (2024). The Dallas Lifespan Brain Study. OpenNeuro. [Dataset] doi: doi:10.18112/openneuro.ds004856.v1.2.0
[15] Peter N. Taylor and Yujiang Wang and Callum Simpson and Vytene Janiukstyte and Jonathan Horsley and Karoline Leiberg and Beth Little and Harry Clifford and Sophie Adler and Sjoerd B. Vos and Gavin P Winston and Andrew W McEvoy and Anna Miserocchi and Jane de Tisi and John S Duncan (2024). The Imaging Database for Epilepsy And Surgery (IDEAS). OpenNeuro. [Dataset] doi: doi:10.18112/openneuro.ds005602.v1.0.0
[16] Makayla Gibson and Roger Newman-Norlund and Leonardo Bonilha and Julius Fridriksson and Gregory Hickok and Argye E. Hillis and Dirk-Bart den Ouden and Chris Rorden (2024). Aphasia Recovery Cohort (ARC) Dataset. OpenNeuro. [Dataset] doi: doi:10.18112/openneuro.ds004884.v1.0.2
[17] David Seminowicz and Shana Burrowes and Alexandra Kearson and Jing Zhang and Samuel Krimmel and Luma Samawi and Andrew Furman and Michael Keaser (2024). MBSR. OpenNeuro. [Dataset] doi: doi:10.18112/openneuro.ds005016.v1.1.1
[18] Bilder, R and Poldrack, R and Cannon, T and London, E and Freimer, N and Congdon, E and Karlsgodt, K and Sabb, F (2018). UCLA Consortium for Neuropsychiatric Phenomics LA5c Study. OpenNeuro. [Dataset] doi:
[19] Strike, Lachlan T. and Hansell, Narelle K. and Miller, Jessica L. and Chuang, Kai-Hsiang and Thompson, Paul M. and de Zubicaray, Greig I. and McMahon, Katie L. and Wright, Margaret J. (2022). Queensland Twin Adolescent Brain (QTAB). OpenNeuro. [Dataset] doi: doi:10.18112/openneuro.ds004146.v1.0.4
[20] Lukas Snoek and Maite van der Miesen and Andries van der Leij and Tinka Beemsterboer and Annemarie Eigenhuis and Steven Scholte (2021). AOMIC-ID1000. OpenNeuro. [Dataset] doi: 10.18112/openneuro.ds003097.v1.2.1
[21] A Munk, J Ambsdorf, S Llambias, M Nielsen (2024). AMAES: Augmented Masked Autoencoder Pretraining on Public Brain MRI Data for 3D-Native Segmentation. In Proceedings of first MICCAI Workshop on Advancing Data Solutions in Medical Imaging AI (ADSMI 2024).
[22] SN Llambias, J Machnio, A Munk, J Ambsdorf, M Nielsen, MM Ghazi (2024). Yucca: A deep learning framework for medical image analysis. ArXiv preprint.
